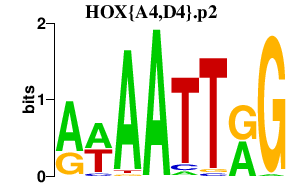
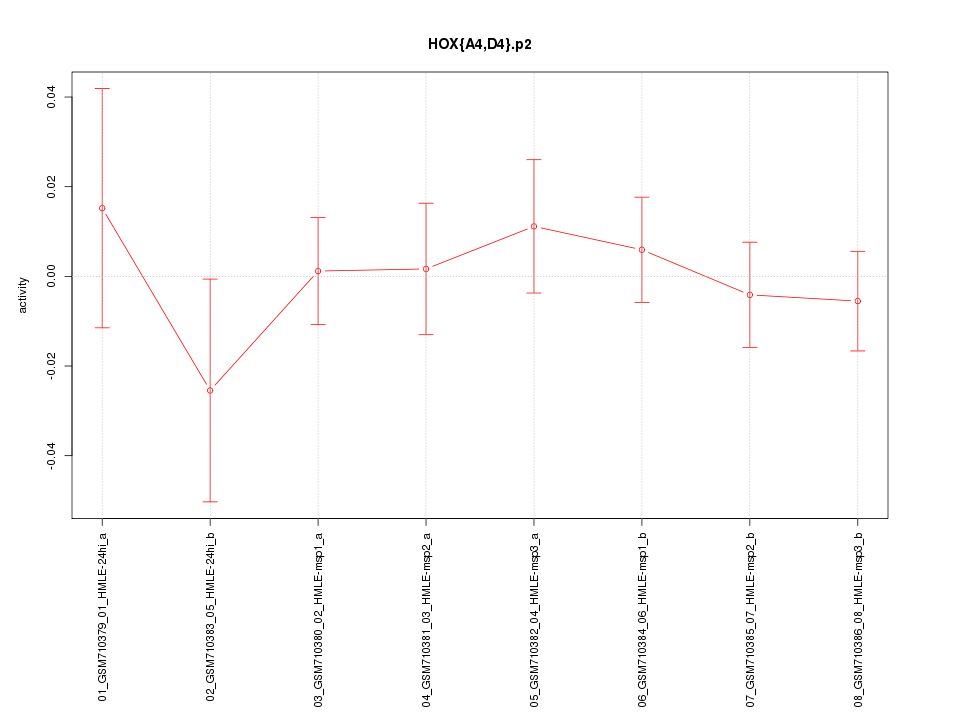
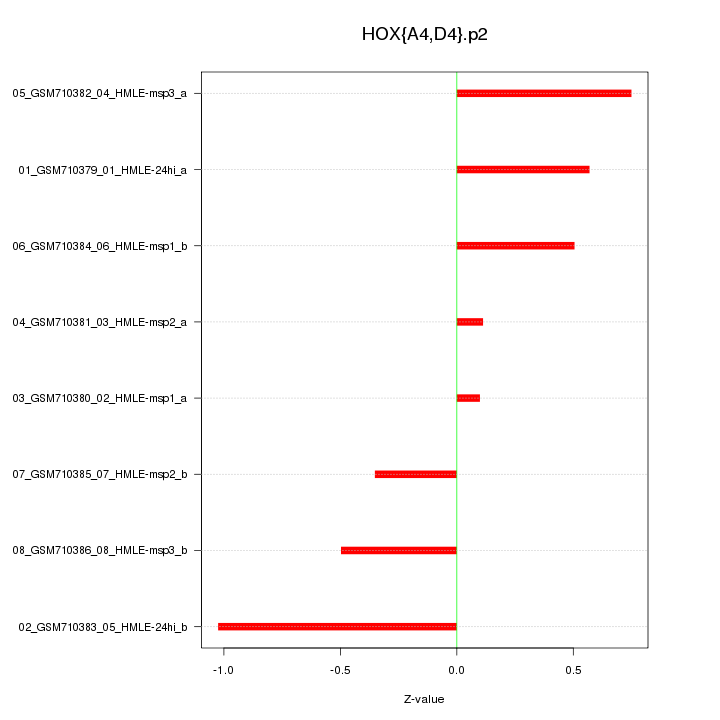
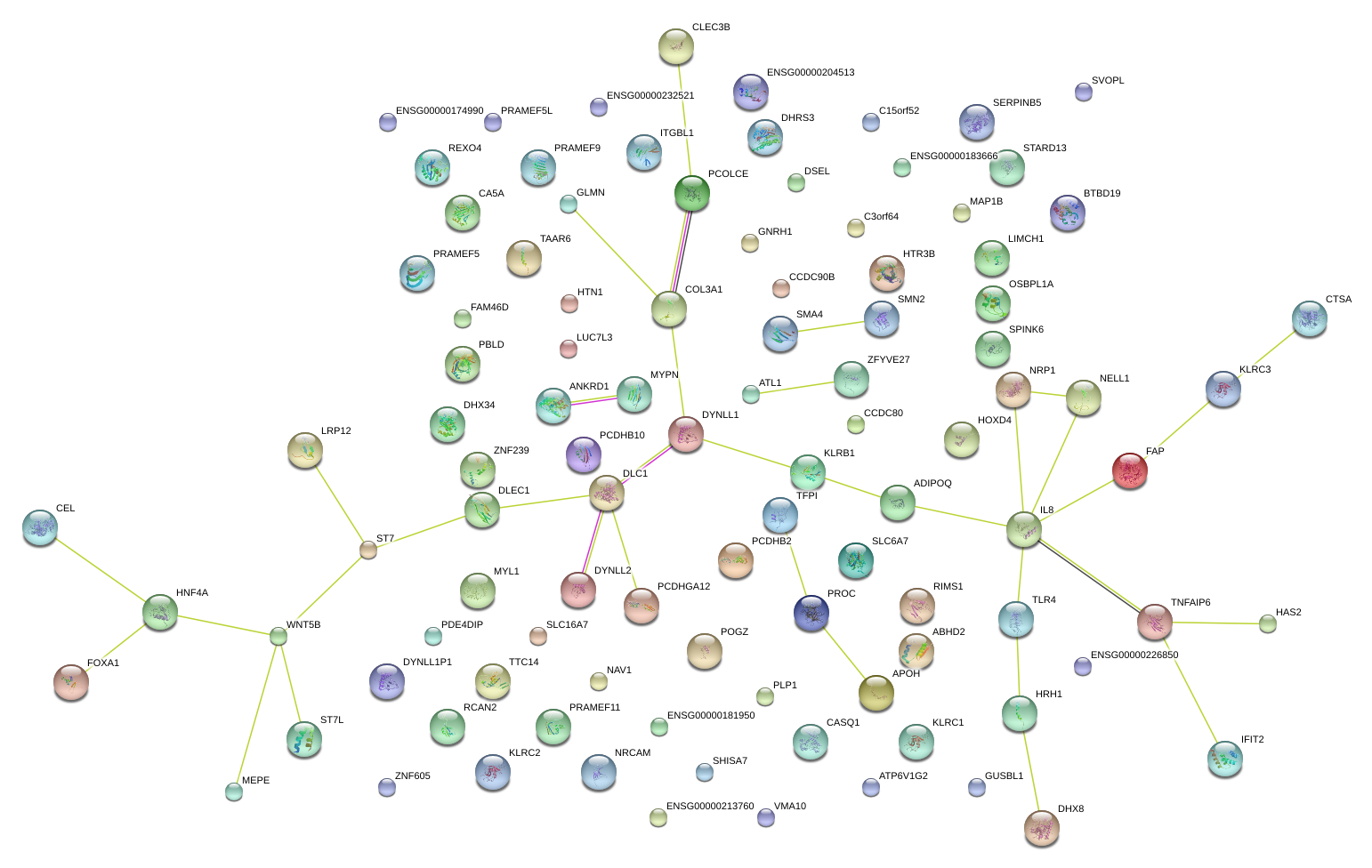
|
chr7_+_100199881
|
0.478
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr6_+_132891460
|
0.459
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr3_+_45067750
|
0.414
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr2_-_163099877
|
0.365
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr10_-_92681025
|
0.327
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_+_127420690
|
0.327
|
|
MIR181A2HG
|
MIR181A2 host gene (non-protein coding)
|
|
chr1_+_160160284
|
0.311
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr12_-_10588543
|
0.310
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr8_-_122653493
|
0.299
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr13_+_102104941
|
0.298
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_+_177016112
|
0.291
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr4_+_70916158
|
0.263
|
NM_002159
|
HTN1
|
histatin 1
|
|
chr12_+_60083125
|
0.262
|
NM_004731
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr10_+_99498233
|
0.260
|
NM_001002261
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr17_+_48821106
|
0.254
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr3_+_180320955
|
0.253
|
|
TTC14
|
tetratricopeptide repeat domain 14
|
|
chr7_-_138363789
|
0.251
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr2_+_128175989
|
0.243
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr2_+_189839078
|
0.242
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_201708648
|
0.238
|
|
NAV1
|
neuron navigator 1
|
|
chr15_-_40630272
|
0.237
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr17_-_64225496
|
0.233
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr12_+_1726221
|
0.226
|
NM_030775
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chrX_+_103031797
|
0.223
|
|
PLP1
|
proteolipid protein 1
|
|
chr18_-_65182507
|
0.218
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr14_+_51026853
|
0.217
|
|
ATL1
|
atlastin GTPase 1
|
|
chr14_+_51026742
|
0.216
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr1_+_45274153
|
0.215
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr10_-_70066594
|
0.214
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr5_+_69696805
|
0.212
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr4_+_74606222
|
0.208
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr3_-_112358510
|
0.200
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr9_+_120466625
|
0.197
|
|
TLR4
|
toll-like receptor 4
|
|
chr8_-_25282555
|
0.197
|
NM_000825
NM_001083111
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone)
|
|
chrX_+_79675945
|
0.195
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr1_-_144997110
|
0.195
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_-_188419049
|
0.194
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_+_177016337
|
0.191
|
|
|
|
|
chr14_-_38064176
|
0.190
|
|
FOXA1
|
forkhead box A1
|
|
chr1_-_12891262
|
0.190
|
NM_001146344
|
PRAMEF11
|
PRAME family member 11
|
|
chr6_-_31514620
|
0.187
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr13_-_33780142
|
0.187
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr20_+_43029973
|
0.186
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr3_-_112359547
|
0.185
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_33623299
|
0.183
|
|
NRP1
|
neuropilin 1
|
|
chr7_-_107880492
|
0.182
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr5_+_149569519
|
0.181
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr8_-_13372183
|
0.180
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_-_69037498
|
0.180
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr22_+_22735202
|
0.180
|
|
|
|
|
chr3_+_11294384
|
0.179
|
NM_000861
|
HRH1
|
histamine receptor H1
|
|
chr6_-_31514393
|
0.179
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr11_+_113775517
|
0.178
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr14_-_38064440
|
0.177
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr3_-_112359565
|
0.177
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr15_+_89631625
|
0.176
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr3_+_186560462
|
0.174
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr18_+_61144143
|
0.174
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr10_+_69869249
|
0.173
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_+_186560478
|
0.172
|
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr10_-_44063906
|
0.172
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr15_+_89631731
|
0.171
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr5_+_140571903
|
0.170
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr6_+_72892371
|
0.170
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr20_+_44518782
|
0.169
|
|
CTSA
|
cathepsin A
|
|
chr10_+_91061705
|
0.169
|
NM_001547
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2
|
|
chrX_+_103031753
|
0.166
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr2_-_188419185
|
0.165
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chrX_+_103031433
|
0.164
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr4_+_71200670
|
0.163
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr3_-_112359251
|
0.159
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_+_189839113
|
0.158
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr5_+_140810181
|
0.156
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr6_-_46293628
|
0.156
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr16_-_87970107
|
0.155
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr1_-_12677347
|
0.154
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr4_+_88754120
|
0.154
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr5_+_140474198
|
0.154
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr15_+_89631695
|
0.154
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr1_-_151431648
|
0.153
|
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr7_+_116660263
|
0.153
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr15_+_89739234
|
0.153
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr5_+_147582338
|
0.153
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr5_+_71403040
|
0.152
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr4_+_41614725
|
0.150
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_+_152214105
|
0.150
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr5_+_140739589
|
0.148
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr2_-_211179765
|
0.148
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr7_-_75443026
|
0.147
|
NM_002991
|
CCL24
|
chemokine (C-C motif) ligand 24
|
|
chr8_-_72268720
|
0.147
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr12_-_29937691
|
0.145
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr3_-_112359277
|
0.145
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_114866090
|
0.144
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_+_97521930
|
0.143
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr7_+_100770378
|
0.143
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr15_-_37391893
|
0.142
|
|
MEIS2
|
Meis homeobox 2
|
|
chr16_+_4421774
|
0.142
|
NM_138440
|
VASN
|
vasorin
|
|
chr5_+_140868721
|
0.141
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr12_-_4754275
|
0.141
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr5_+_140810126
|
0.141
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr7_-_92747269
|
0.140
|
|
SAMD9
|
sterile alpha motif domain containing 9
|
|
chr17_+_53344966
|
0.140
|
|
HLF
|
hepatic leukemia factor
|
|
chr18_+_3247825
|
0.139
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr7_+_116660291
|
0.139
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_+_104097321
|
0.138
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr5_-_96149804
|
0.137
|
NM_001198541
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr7_+_2557496
|
0.136
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr14_+_64680858
|
0.135
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr2_-_32490688
|
0.134
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr12_+_75874512
|
0.132
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr5_+_140810326
|
0.131
|
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr12_+_41831484
|
0.131
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr5_+_67586466
|
0.130
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_+_47624222
|
0.130
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr2_+_128176019
|
0.129
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr17_-_39092714
|
0.129
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr10_-_92680784
|
0.129
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr8_-_108510094
|
0.128
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr12_-_95467104
|
0.128
|
NM_001032287
NM_001127362
NM_003297
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1
|
|
chr1_+_150245182
|
0.128
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr17_+_7384720
|
0.127
|
NM_001102614
|
SLC35G6
|
solute carrier family 35, member G6
|
|
chr17_-_53809444
|
0.127
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr7_+_141811508
|
0.126
|
|
LOC93432
|
maltase-glucoamylase (alpha-glucosidase) pseudogene
|
|
chr1_+_215178884
|
0.125
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr3_+_108541544
|
0.125
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr3_-_52737599
|
0.124
|
NM_152932
|
GLT8D1
|
glycosyltransferase 8 domain containing 1
|
|
chr17_-_67138013
|
0.123
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr8_+_38831667
|
0.122
|
NM_153692
|
HTRA4
|
HtrA serine peptidase 4
|
|
chr3_+_46619075
|
0.122
|
NM_003212
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr3_-_178984789
|
0.122
|
NM_001163677
NM_171828
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr16_+_55515468
|
0.121
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr7_+_23286384
|
0.119
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_+_161676761
|
0.119
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr9_-_8733893
|
0.119
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr8_+_1952875
|
0.119
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr2_+_189839132
|
0.118
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr17_-_38911524
|
0.118
|
NM_181534
|
KRT25
|
keratin 25
|
|
chr1_+_201709051
|
0.117
|
|
NAV1
|
neuron navigator 1
|
|
chr6_+_29274402
|
0.116
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chr3_-_74570262
|
0.116
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr19_-_12845539
|
0.115
|
|
C19orf43
|
chromosome 19 open reading frame 43
|
|
chr3_+_130569368
|
0.115
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr10_-_69455926
|
0.114
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr17_-_38721701
|
0.114
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chrX_+_144902865
|
0.114
|
NM_001144006
NM_001144008
NM_001144009
NM_001144010
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chrY_-_19882311
|
0.113
|
NM_001002906
NM_004677
|
XKRY2
XKRY
|
XK, Kell blood group complex subunit-related, Y-linked 2
XK, Kell blood group complex subunit-related, Y-linked
|
|
chr14_+_21756135
|
0.113
|
NM_020366
|
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1
|
|
chr5_-_138210992
|
0.113
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chrX_-_112066353
|
0.112
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr12_-_10605928
|
0.111
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr20_-_4990913
|
0.111
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr4_-_41750936
|
0.111
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr1_+_40735749
|
0.110
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chrM_+_4465
|
0.109
|
|
|
|
|
chrM_+_6138
|
0.109
|
|
|
|
|
chr4_-_156787377
|
0.108
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr22_+_31366662
|
0.108
|
|
|
|
|
chr3_+_73110809
|
0.108
|
NM_018029
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2
|
|
chr3_-_18480203
|
0.107
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr3_+_69134057
|
0.107
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr5_+_126988706
|
0.106
|
NM_001127385
|
CTXN3
|
cortexin 3
|
|
chr2_-_96482382
|
0.106
|
|
LINC00342
|
long intergenic non-protein coding RNA 342
|
|
chr20_-_17641122
|
0.105
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr17_-_79269082
|
0.104
|
NM_001037984
NM_138570
|
SLC38A10
|
solute carrier family 38, member 10
|
|
chr9_-_27005649
|
0.104
|
NM_022901
|
LRRC19
|
leucine rich repeat containing 19
|
|
chr3_-_48601200
|
0.103
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr8_-_139926235
|
0.103
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chrX_+_16964730
|
0.103
|
NM_001080975
NM_004726
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr7_+_23286390
|
0.102
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr5_+_156696351
|
0.102
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr11_-_71752099
|
0.102
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr3_-_114343052
|
0.101
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_+_72658735
|
0.100
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr7_-_126883568
|
0.100
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr17_+_3379295
|
0.100
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr3_-_47933997
|
0.098
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr12_+_15699277
|
0.098
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr11_-_94134575
|
0.098
|
NM_016540
|
GPR83
|
G protein-coupled receptor 83
|
|
chr2_+_87144737
|
0.098
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr20_+_61476475
|
0.098
|
NM_080750
|
DPH3P1
|
DPH3, KTI11 homolog (S. cerevisiae) pseudogene 1
|
|
chr5_+_140430960
|
0.097
|
NM_013340
|
PCDHB1
|
protocadherin beta 1
|
|
chr1_+_174843523
|
0.097
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chrX_+_99899162
|
0.096
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr5_-_138211054
|
0.096
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr19_+_4153593
|
0.096
|
NM_032607
|
CREB3L3
|
cAMP responsive element binding protein 3-like 3
|
|
chr11_+_22688156
|
0.096
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr3_+_97868169
|
0.096
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr16_+_31366468
|
0.095
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr12_+_56522049
|
0.095
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr3_-_114102488
|
0.095
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr16_-_73092533
|
0.095
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr17_-_39538635
|
0.094
|
NM_021013
|
KRT34
|
keratin 34
|
|
chr17_+_30677187
|
0.094
|
|
ZNF207
|
zinc finger protein 207
|
|
chr16_+_24740945
|
0.094
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr13_-_24007822
|
0.093
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr22_+_39353526
|
0.093
|
NM_001193289
NM_145699
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A
|
|
chr2_+_183582667
|
0.093
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr13_+_78109808
|
0.093
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr12_+_56522043
|
0.092
|
|
ESYT1
|
extended synaptotagmin-like protein 1
|
|
chr5_-_41510627
|
0.092
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|